publications
publications by categories in reversed chronological order. generated by jekyll-scholar.
2024
-
 Molecular underpinnings of plasticity and supergene-mediated polymorphism in fire ant queensAlex H. Waugh, Michael A. Catto, Samuel V. Arsenault, Sasha Kay, Kenneth G. Ross, and Brendan G. HuntJournal of Evolutionary Biology 2024
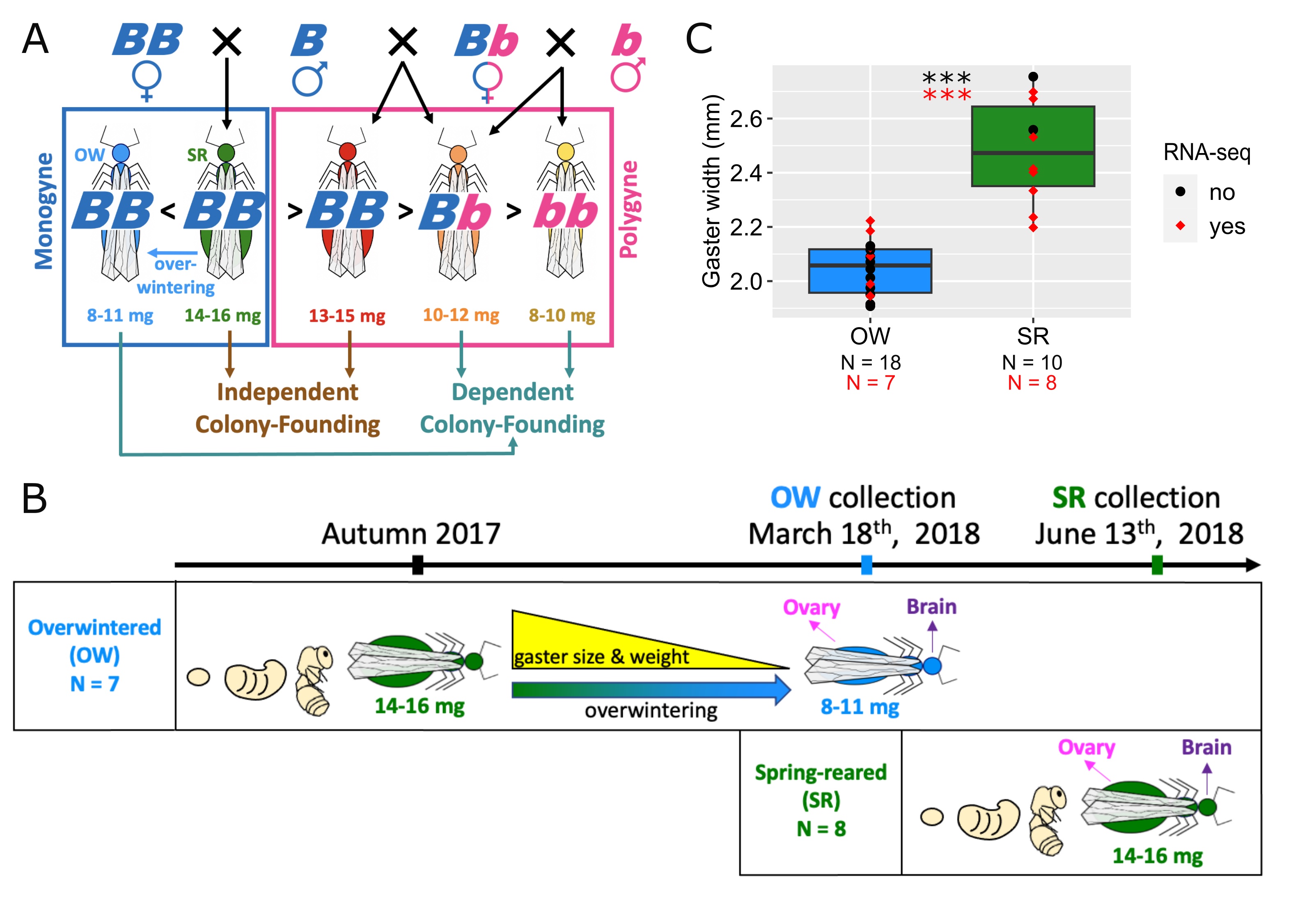
Molecular underpinnings of plasticity and supergene-mediated polymorphism in fire ant queensAlex H. Waugh, Michael A. Catto, Samuel V. Arsenault, Sasha Kay, Kenneth G. Ross, and Brendan G. HuntJournal of Evolutionary Biology 2024Characterizing molecular underpinnings of plastic traits and balanced polymorphisms represent two important goals of evolutionary biology. Fire ant gynes (pre-reproductive queens) provide an ideal system to study potential links between these phenomena because they exhibit both supergene-mediated polymorphism and nutritional plasticity in weight and colony-founding behavior. Gynes with the inversion supergene haplotype are lightweight and depend on existing workers to initiate reproduction. Gynes with only the ancestral, non-inverted gene arrangement accumulate more nutrient reserves as adults and, in a distinct colony-founding behavior, initiate reproduction without help from workers. However, when such gynes overwinter in the natal nest they develop an environmentally induced lightweight phenotype and colony-founding behavior, similar to gynes with the inversion haplotype that have not overwintered. To evaluate the extent of shared mechanisms between plasticity and balanced polymorphism in fire ant gyne traits, we assessed whether genes with expression variation linked to overwintering plasticity may be affected by evolutionary divergence between supergene haplotypes. To do so, we first compared transcriptional profiles of brains and ovaries from overwintered and non-overwintered gynes to identify plasticity-associated genes. These genes were enriched for metabolic and behavioral functions. Next, we compared plasticity-associated genes to those differentially expressed by supergene genotype, revealing a significant overlap of the two sets in ovarian tissues. We also identified sequence substitutions between supergene variants of multiple plasticity-associated genes, consistent with a scenario in which an ancestrally plastic phenotype responsive to an environmental condition became increasingly genetically regulated.
2023
-
 A caste differentiation mutant elucidates the evolution of socially parasitic antsWaring Trible, Vikram Chandra, Kip D. Lacy, Gina Limón, Sean K. McKenzie, Leonora Olivos-Cisneros, Samuel V. Arsenault, and Daniel J.C. KronauerCurrent Biology 2023
A caste differentiation mutant elucidates the evolution of socially parasitic antsWaring Trible, Vikram Chandra, Kip D. Lacy, Gina Limón, Sean K. McKenzie, Leonora Olivos-Cisneros, Samuel V. Arsenault, and Daniel J.C. KronauerCurrent Biology 2023Most ant species have two distinct female castes—queens and workers—yet the developmental and genetic mechanisms that produce these alternative phenotypes remain poorly understood. Working with a clonal ant, we discovered a variant strain that expresses queen-like traits in individuals that would normally become workers. The variants show changes in morphology, behavior, and fitness that cause them to rely on workers in wild-type (WT) colonies for survival. Overall, they resemble the queens of many obligately parasitic ants that have evolutionarily lost the worker caste and live inside colonies of closely related hosts. The prevailing theory for the evolution of these workerless social parasites is that they evolve from reproductively isolated populations of facultative intermediates that acquire parasitic phenotypes in a stepwise fashion. However, empirical evidence for such facultative ancestors remains weak, and it is unclear how reproductive isolation could gradually arise in sympatry. In contrast, we isolated these variants just a few generations after they arose within their WT parent colony, implying that the complex phenotype reported here was induced in a single genetic step. This suggests that a single genetic module can decouple the coordinated mechanisms of caste development, allowing an obligately parasitic variant to arise directly from a free-living ancestor. Consistent with this hypothesis, the variants have lost one of the two alleles of a putative supergene that is heterozygous in WTs. These findings provide a plausible explanation for the evolution of ant social parasites and implicate new candidate molecular mechanisms for ant caste differentiation.
2022
-
 Direct and indirect genetic effects of a social supergeneSamuel V. Arsenault, Oksana Riba-Grognuz, DeWayne Shoemaker, Brendan G. Hunt, and Laurent KellerMolecular Ecology 2022
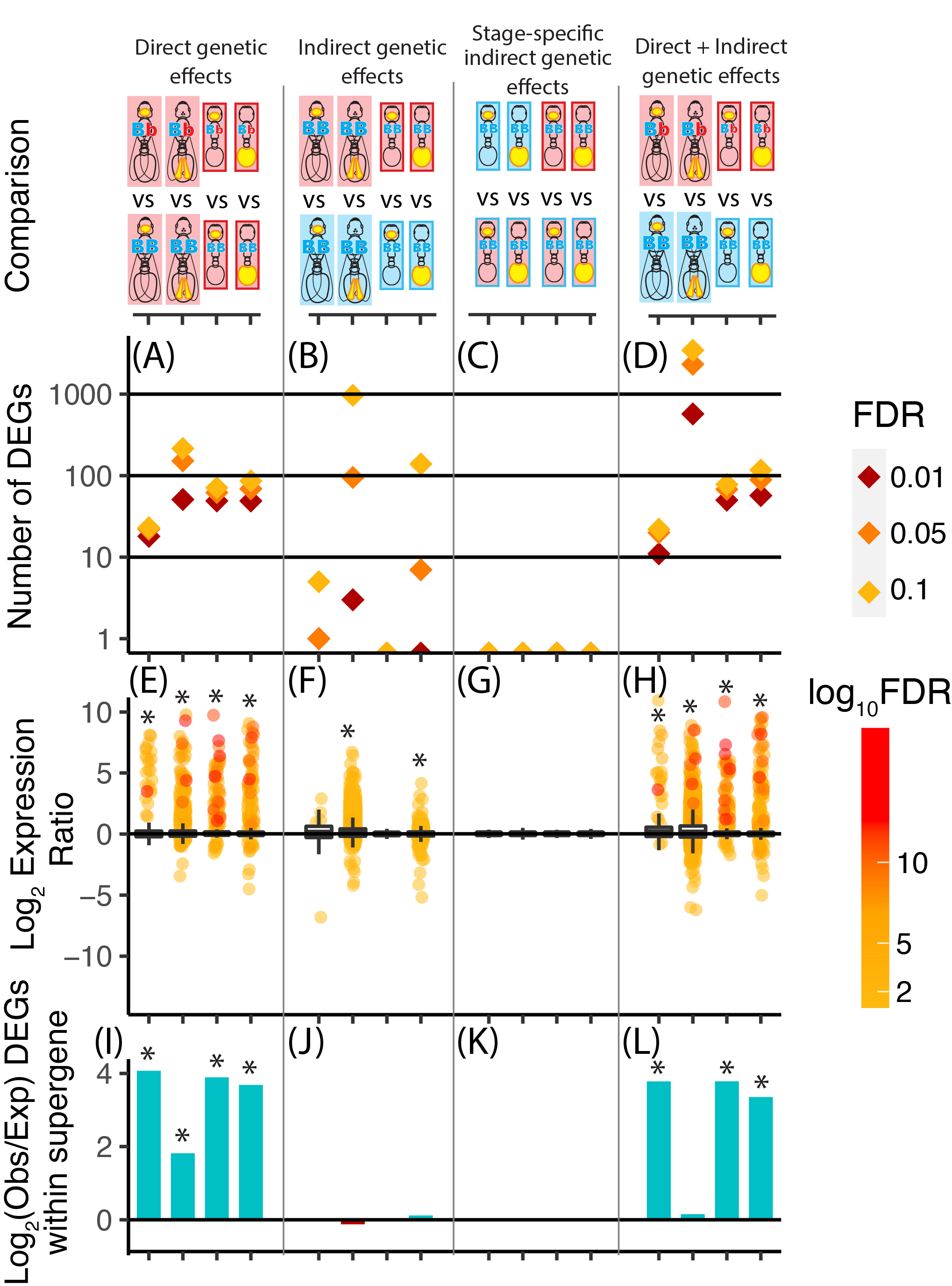
Direct and indirect genetic effects of a social supergeneSamuel V. Arsenault, Oksana Riba-Grognuz, DeWayne Shoemaker, Brendan G. Hunt, and Laurent KellerMolecular Ecology 2022Indirect genetic effects describe phenotypic variation that results from differences in the genotypic composition of social partners. Such effects represent heritable sources of environmental variation in eusocial organisms because individuals are typically reared by their siblings. In the fire ant Solenopsis invicta, a social supergene exhibits striking indirect genetic effects on worker regulation of colony queen number, such that the genotypic composition of workers at the supergene determines whether colonies contain a single or multiple queens. We assessed the direct and indirect genetic effects of this supergene on gene expression in brains and abdominal tissues from lab-reared workers and compared these with previously published data from field-collected pre-reproductive queens. We found that direct genetic effects caused larger gene expression changes and were more consistent across tissue types and castes than indirect genetic effects. Indirect genetic effects influenced the expression of many loci but were generally restricted to the abdominal tissues. Further, indirect genetic effects were only detected when the genotypic composition of social partners differed throughout the development and adult life of focal workers, and were often only significant with relatively lenient statistical cutoffs. Our study provides insight into direct and indirect genetic effects of a social supergene on gene regulatory dynamics across tissues and castes in a complex society.
2021
2020
-
 Simple inheritance, complex regulation: Supergene‐mediated fire ant queen polymorphismSamuel V. Arsenault, Joanie T. King, Sasha Kay, Kip D. Lacy, Kenneth G. Ross, and Brendan G. HuntMolecular Ecology Oct 2020
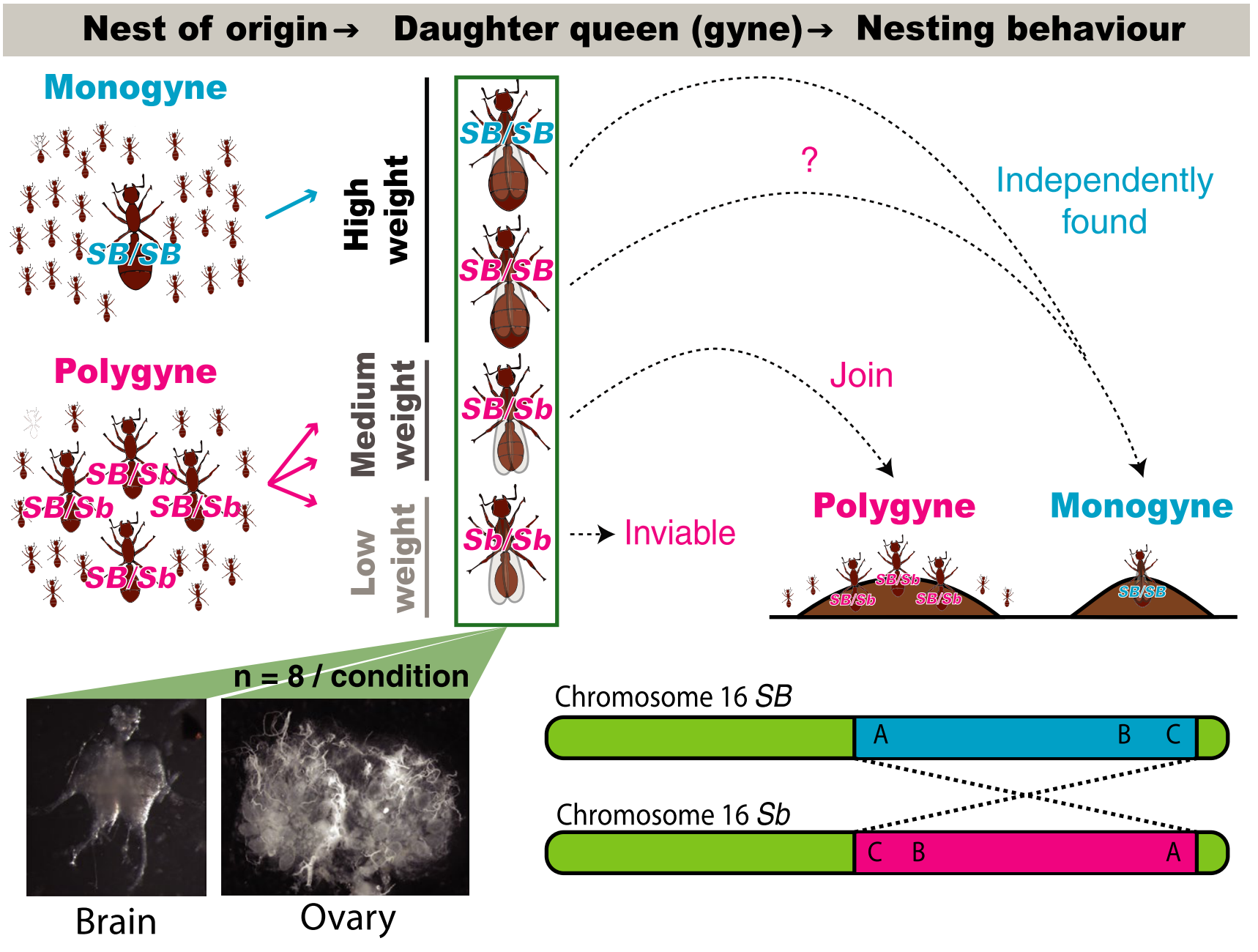
Simple inheritance, complex regulation: Supergene‐mediated fire ant queen polymorphismSamuel V. Arsenault, Joanie T. King, Sasha Kay, Kip D. Lacy, Kenneth G. Ross, and Brendan G. HuntMolecular Ecology Oct 2020The fire ant Solenopsis invicta exists in two alternate social forms: monogyne nests contain a single reproductive queen and polygyne nests contain multiple reproduc- tive queens. This colony-level social polymorphism corresponds with individual dif- ferences in queen physiology, queen dispersal patterns and worker discrimination behaviours, all evidently regulated by an inversion-based supergene that spans more than 13 Mb of a “social chromosome,” contains over 400 protein-coding genes and rarely undergoes recombination. The specific mechanisms by which this supergene influences expression of the many distinctive features that characterize the alternate forms remain almost wholly unknown. To advance our understanding of these mech- anisms, we explore the effects of social chromosome genotype and natal colony so- cial form on gene expression in queens sampled as they embarked on nuptial flights, using RNA-sequencing of brains and ovaries. We observe a large effect of natal social form, that is, of the social/developmental environment, on gene expression profiles, with similarly substantial effects of genotype, including: (a) supergene-associated gene upregulation, (b) allele-specific expression and (c) pronounced extra-supergene trans-regulatory effects. These findings, along with observed spatial variation in dif- ferential and allele-specific expression within the supergene region, highlight the complex gene regulatory landscape that emerged following divergence of the inver- sion-mediated Sb haplotype from its homologue, which presumably largely retained the ancestral gene order. The distinctive supergene-associated gene expression tra- jectories we document at the onset of a queen’s reproductive life expand the known record of relevant molecular correlates of a complex social polymorphism and point to putative genetic factors underpinning the alternate social syndromes.
-
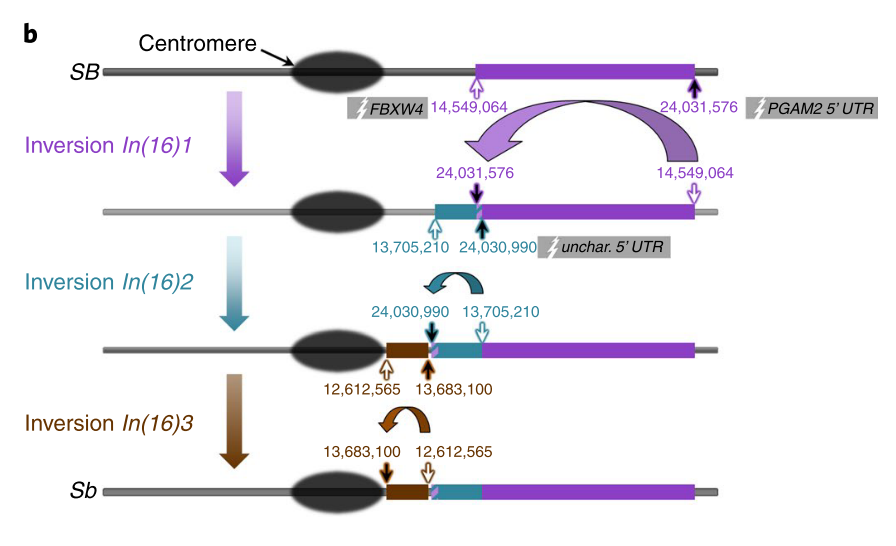 Evolution of a supergene that regulates a trans-species social polymorphismZheng Yan, Simon H. Martin, Dietrich Gotzek, Samuel V. Arsenault, Pablo Duchen, Quentin Helleu, Oksana Riba-Grognuz, Brendan G. Hunt, Nicolas Salamin, Dewayne Shoemaker, and 2 more authorsNature Ecology & Evolution Feb 2020
Evolution of a supergene that regulates a trans-species social polymorphismZheng Yan, Simon H. Martin, Dietrich Gotzek, Samuel V. Arsenault, Pablo Duchen, Quentin Helleu, Oksana Riba-Grognuz, Brendan G. Hunt, Nicolas Salamin, Dewayne Shoemaker, and 2 more authorsNature Ecology & Evolution Feb 2020Supergenes are clusters of linked genetic loci that jointly affect the expression of complex phenotypes, such as social organization. Little is known about the origin and evolution of these intriguing genomic elements. Here we analyse whole-genome sequences of males from native populations of six fire ant species and show that variation in social organization is under the control of a novel supergene haplotype (termed Sb), which evolved by sequential incorporation of three inversions spanning half of a ‘social chromosome’. Two of the inversions interrupt protein-coding genes, resulting in the increased expression of one gene and modest truncation in the primary protein structure of another. All six socially polymorphic species studied harbour the same three inversions, with the single origin of the supergene in their common ancestor inferred by phylogenomic analyses to have occurred half a million years ago. The persistence of Sb along with the ancestral SB haplotype through multiple speciation events provides a striking example of a functionally important trans-species social polymorphism presumably maintained by balancing selection. We found that while recombination between the Sb and SB haplotypes is severely restricted in all species, a low level of gene flux between the haplotypes has occurred following the appearance of the inversions, potentially mitigating the evolutionary degeneration expected at genomic regions that cannot freely recombine. These results provide a detailed picture of the structural genomic innovations involved in the formation of a supergene controlling a complex social phenotype.
2019
- Single Cells of Neurospora Crassa Show Circadian Oscillations, Light Entrainment, Temperature Compensation, and Phase SynchronizationZhaojie Deng, Jia Hwei Cheong, Cristian Caranica, Lingyun Wu, Xiao Qiu, Michael T. Judge, Brooke Hull, Carmen Rodriguez, James Griffith, Ahmad Al-Omari, and 4 more authorsIEEE Access Feb 2019
-
 Leveraging technological innovations to investigate evolutionary transitions to eusocialitySamuel V. Arsenault, Karl M. Glastad, and Brendan G. HuntCurrent Opinion in Insect Science Feb 2019
Leveraging technological innovations to investigate evolutionary transitions to eusocialitySamuel V. Arsenault, Karl M. Glastad, and Brendan G. HuntCurrent Opinion in Insect Science Feb 2019The study of the major transition to eusociality presents several challenges to researchers, largely resulting from the importance of complex behavioral phenotypes and the shift from individual to group level selection. These challenges are being met with corresponding technological improvements. Advances in resource development for non-model taxa, behavioral tracking, nucleic acid sequencing, and reverse genetics are facilitating studies of hypotheses that were previously intractable. These innovations are resulting in the development of new model systems tailored to the exploration of specific behavioral phenotypes and the querying of underlying molecular mechanisms that drive eusocial behaviors. Here, we present a brief overview of how methodological innovations are advancing our understanding of the evolution of eusociality.
2018
-
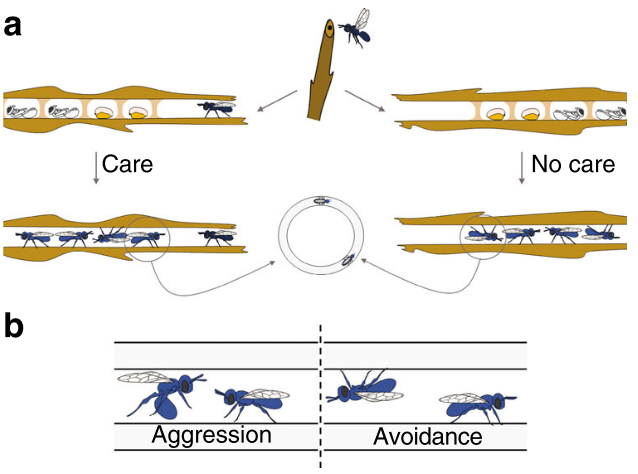 The effect of maternal care on gene expression and DNA methylation in a subsocial beeSamuel V. Arsenault, Brendan G. Hunt, and Sandra M. RehanNature Communications Dec 2018
The effect of maternal care on gene expression and DNA methylation in a subsocial beeSamuel V. Arsenault, Brendan G. Hunt, and Sandra M. RehanNature Communications Dec 2018Developmental plasticity describes the influence of environmental factors on phenotypic variation. An important mediator of developmental plasticity in many animals is parental care. Here, we examine the consequences of maternal care on offspring after the initial mass provisioning of brood in the small carpenter bee, Ceratina calcarata. Removal of the mother during larval development leads to increased aggression and avoidance in adulthood. This corresponds with changes in expression of over one thousand genes, alternative splicing of hundreds of genes, and significant changes to DNA methylation. We identify genes related to metabolic and neuronal functions that may influence developmental plasticity and aggression. We observe no genome-wide association between differential DNA methylation and differential gene expression or splicing, though indirect relationships may exist between these factors. Our results provide insight into the gene regulatory context of DNA methylation in insects and the molecular avenues through which variation in maternal care influences developmental plasticity.
2017
-
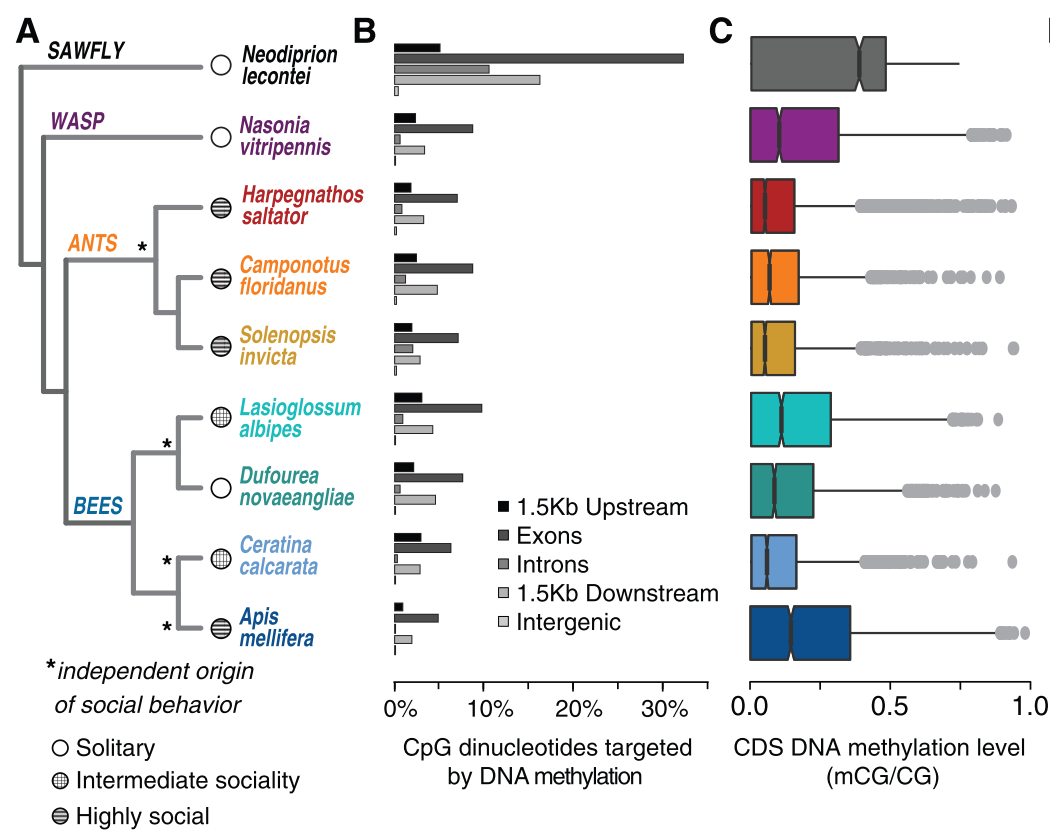 Variation in DNA Methylation Is Not Consistently Reflected by Sociality in HymenopteraKarl M. Glastad, Samuel V. Arsenault, Kim L. Vertacnik, Scott M. Geib, Sasha Kay, Bryan N. Danforth, Sandra M. Rehan, Catherine R. Linnen, Sarah D. Kocher, and Brendan G. HuntGenome Biology and Evolution Jun 2017
Variation in DNA Methylation Is Not Consistently Reflected by Sociality in HymenopteraKarl M. Glastad, Samuel V. Arsenault, Kim L. Vertacnik, Scott M. Geib, Sasha Kay, Bryan N. Danforth, Sandra M. Rehan, Catherine R. Linnen, Sarah D. Kocher, and Brendan G. HuntGenome Biology and Evolution Jun 2017Changes in gene regulation that underlie phenotypic evolution can be encoded directly in the DNA sequence or mediated by chromatin modifications such as DNA methylation. It has been hypothesized that the evolution of eusocial division of labor is associated with enhanced gene regulatory potential, which may include expansions in DNA methylation in the genomes of Hymenoptera (bees, ants, wasps, and sawflies). Recently, this hypothesis garnered support from analyses of a commonly used metric to estimate DNA methylation in silico, CpG content. Here, we test this hypothesis using direct, nucleotide-level measures of DNAmethylationacross ninespeciesofHymenoptera. In doing so,wegeneratednewDNAmethylomes for three speciesof interest, including one solitary and one facultatively eusocial halictid bee and a sawfly. We demonstrate that the strength of correlation between CpG content and DNA methylation varies widely among hymenopteran taxa, highlighting shortcomings in the utility of CpG content as a proxy for DNA methylation in comparative studies of taxa with sparse DNAmethylomes. We observed strikingly high levels of DNAmethylation in the sawfly relative to other investigated hymenopterans. Analyses of molecular evolution suggest the relatively distinct sawfly DNAmethylome may be associatedwith positive selection on functional DNMT3domains. Sawflies are an outgroup to all ants, bees, and wasps, and no sawfly species are eusocial. We find no evidence that either global expansions or variation within individual ortholog groups in DNA methylation are consistently associated with the evolution of social behavior.
2016
-
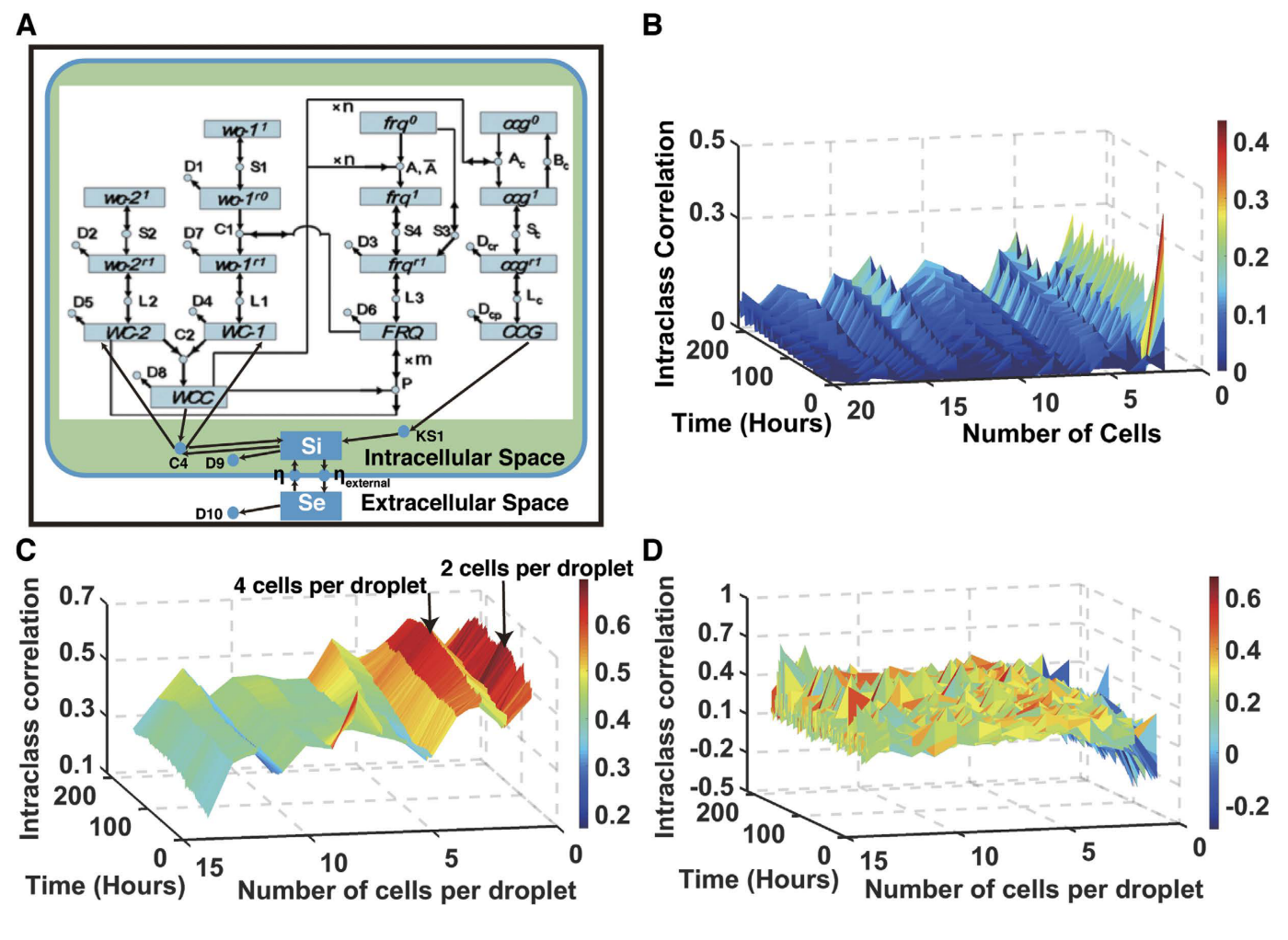 Synchronizing stochastic circadian oscillators in single cells of Neurospora crassaZhaojie Deng, Samuel V. Arsenault, Cristian Caranica, James Griffith, Taotao Zhu, Ahmad Al-Omari, Heinz-Bernd Schuttler, Jonathan Arnold, and Leidong MaoScientific Reports Jun 2016
Synchronizing stochastic circadian oscillators in single cells of Neurospora crassaZhaojie Deng, Samuel V. Arsenault, Cristian Caranica, James Griffith, Taotao Zhu, Ahmad Al-Omari, Heinz-Bernd Schuttler, Jonathan Arnold, and Leidong MaoScientific Reports Jun 2016The synchronization of stochastic coupled oscillators is a central problem in physics and an emerging problem in biology, particularly in the context of circadian rhythms. Most measurements on the biological clock are made at the macroscopic level of millions of cells. Here measurements are made on the oscillators in single cells of the model fungal system, Neurospora crassa, with droplet microfluidics and the use of a fluorescent recorder hooked up to a promoter on a clock controlled gene-2 (ccg-2). The oscillators of individual cells are stochastic with a period near 21 hours (h), and using a stochastic clock network ensemble fitted by Markov Chain Monte Carlo implemented on general-purpose graphical processing units (or GPGPUs) we estimated that >94% of the variation in ccg-2 expression was stochastic (as opposed to experimental error). To overcome this stochasticity at the macroscopic level, cells must synchronize their oscillators. Using a classic measure of similarity in cell trajectories within droplets, the intraclass correlation (ICC), the synchronization surface ICC is measured on >25,000 cells as a function of the number of neighboring cells within a droplet and of time. The synchronization surface provides evidence that cells communicate, and synchronization varies with genotype.
2014
- Single cell measurements on the biological clock by microfluidicsZhaojie Deng, Samuel V. Arsenault, Taotao Zhu, Rui Cheng, James Griffith, Jonathan Arnold, and Leidong MaoProceedings of the 18th International Conference on Miniaturized Systems for Chemistry and Life Sciences Jun 2014
This paper reports a droplet microfluidic system that is capable of generating droplets, encapsulating cells, incubating cells, and measuring biological clock behavior via fluorescent signals on a large number of single cells simultaneously over a long period of time ( 48 hours).